What is a Domain-Specific Language? It is more common than you think.
Few of us – computational chemists – have heard the term Domain-Specific Language (DSL for short), even though this is somewhat ubiquitous in our workflow, especially when dealing with molecule visualisation. Don’t you believe me? Check out the command you type on VMD, PyMOL, YASARA, or Chimera.
YASARA command:
LoadPDB 1crn, download=yes
AutoPosOriObj 1, x = 1, y = 10, z = 0, alpha = 0, beta = 0, gamma = 0
ColorRes cys, green
ShowRes cys
BallStickRes cys
AutoRotateObj 1, Y = 1
wait 180
AutoRotateObj 1, Y = 0
ZoomRes Cys 16, step = 50
wait 100
ZoomRes all
VMD Tcl/Tk command:
set crystal [atomselect top "all"]
$crystal moveby {10 0 0}
$crystal move [transaxis x 40 deg]
set sel [atomselect top "hydrophobic and alpha"]
$sel get resname
$sel get resid
$sel get {resname resid}
$sel get {x y z}
graphics top cylinder {15 0 0} {15 0 10} radius 10 resolution 60 filled no
graphics top cylinder {0 0 0} {-15 0 10} radius 5 resolution 60 filled yes
graphics top cone {40 0 0} {40 0 10} radius 10 resolution 60
graphics top sphere {65 0 5} radius 10 resolution 80
graphics top triangle {80 0 0} {85 0 10} {90 0 0}
graphics top text {40 0 20} "my drawing objects"
PyMOL command:
select active, (resi 14-20,38) and chain A
zoom active
hide all
show stick, active
select active_water, ( (resi 38 and name ND2 and chain A) around 3.5) and (resn HOH)
show spheres, active_water
alter active_water, vdw=0.5
rebuild
isomesh mesh1, 2fofc.map, 1.0, (resi 14-20,38 and chain A), carve=1.6
isomesh mesh1, 2fofc.map, 1.0, active, carve=1.6
color grey, mesh1
bg_color white
As you can see, DSL is a programming language that is… well, as it says, dealing with a specific domain. For example, the DSL used by YASARA, PyMOL, and VMD is for molecular file I/O processing, molecule selection, manipulation, and visualisation. In other words, those DSLs are specifically targeting molecular information, model, and visualisation domain.
Some other well-known DSLs include R, Matlab, SQL, HTML&CSS, and spreadsheet formulas, which correspond to statistics, modeling, database, webpage, and spreadsheet domain.
But then, what are the differences between General Programming Languages (GPLs) like Java, C++, Fortran, or Python compared to DSL? Also, wouldn’t it be too much hassle for us to learn a programming language to solve a specific problem? Compared to GPLs, DSL is far simpler because DSL uses far more limited vocabulary, grammar, and syntax. Using vocabulary specific to a particular domain alone is already lowering the learning barrier significantly since the domain expert (e.g., computational chemist) is familiar with the DSL commands (verb or noun). And thus, the domain expert could correlate most of the commands with their mental model. For example, even when a computational chemist never used VMD before, (s)he will immediately understand what protein and within 4.5 of resname TPF means.
Well, of course, since the commands and operations in DSL are not as rich as GPL, wouldn’t that make DSL less expressive than GPL? Ideally, the DSL should be expressive enough to allow the domain expert to solve every problem that (s)he faces in a specific domain. Therefore DSL should be as simple as possible yet still allow the domain expert to express the solution to their problem.
Voelter, in his book DSL Engineering (2013), explains that one should attempt to make DSL as close as possible to its target domain (Figure 1). Also, ideally, the DSL should not be under-approximated, in which case it lacks the capability to express the solution in its target domain. On the other hand, it should not over-approximate where it is way too complicated (or overengineered) so that it is capable of doing the operation or implementation that is not required, which is somewhat similar to violating the YAGNI principle (Figure 2).
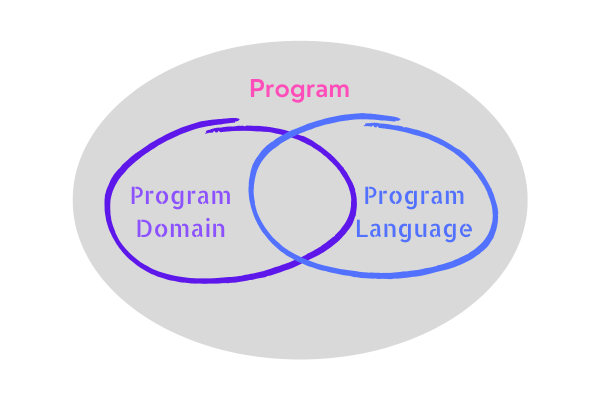
Figure 1. Program Domain and Program Language are subsets of Program sets. DSL (program language) is supposed to overlap as much as possible with its target domain.
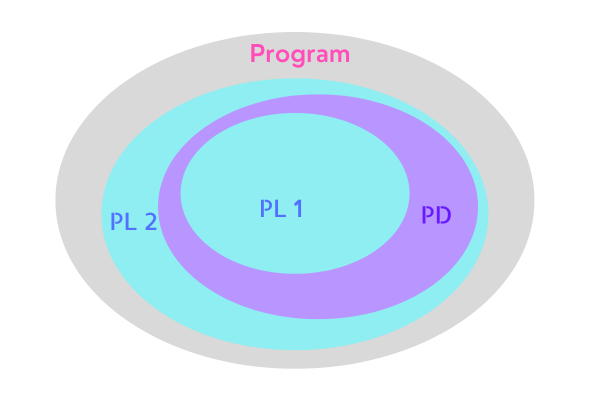
Figure 2. PL 1 is Program Language 1 (DSL 1) and PL 2 is Program Language 2 (DSL 2), PL 1 is under-approximated while PL 2 is over-approximated.
Overall, when DSL is carefully designed and implemented, it can be a powerful yet concise tool for domain experts to express the solution in the target domain.
I believe that is enough explanation for DSL. I choose to explain DSL first so that I can explain to you why we need DSL for molecular interaction analysis in the next section.
Is it really necessary to bring DSL to molecular interaction analysis?
As someone who has been dealing with interaction fingerprinting for more than a decade, I am fully aware of the severe limitation of the solution that can be offered by an Interaction Fingerprinting (IFP) tool.
For example, the IFP tools are usually running according to the choices specified in the configuration file. In other cases, it uses a simple form or wizard using GUI or web interface. This way, the path that can be taken is very limited because:
- The developers expect the users to take only the happy path so that nothing can go wrong.
- Many things have been taken care of under the hood so that the users do not have to deal with so many technical-level details.
- Integrating a new solution into the existing software is difficult. For example, in electrostatic interaction analysis, to modify the atom typing algorithm for positively charged atoms, you have to change the code or SMARTS pattern that identifies the positively charged atoms. One way to overcome this is by providing a plugin system that allows the user to integrate a new solution into the IFP tool. FingeRNAt uses this approach to allow user-defined interaction.
On the other end of the spectrum, there is ProLIF, a Python library for molecular interaction analysis. This means that rather than using a command line + configuration file or graphical interface, it uses a series of commands that will be executed. The benefit of this approach is the expressiveness of this tool, as it gives the user more control starting from which molecular fragment to analyze, the result from presentation for reporting or further analysis, adding or changing the geometry rule for a particular interaction, and of course defining new scoring function for interaction similarity.
The only caveat is, first of all, you should be able to use Python language to a certain degree of mastery. Second, if you are doing an edge case study, there is a chance you have to extend the deeper level implementation in ProLIF which IMO is not trivial.
Also, although in theory, it is possible to express every program domain using Python, ProLIF, and RDKit (as one of ProLIF dependencies), in practice trying to implement a program domain that is not implemented by the ProLIF library will not be easy. For example, we can observe in the ProLIF community that few demanded ProLIF be able to present the interaction result at the atomic level. And to overcome this limitation, the ProLIF developer has to change the core source code so that the interactions are not only registered at the residue level but also at the atomic level.
Whichever the tools that computational chemist use, it seems like there is no end to the creativity and expressiveness in interaction fingerprinting. The feature demands for IFP tools are incredible, starting from support for various file formats, geometric criteria for each interaction, atom typing, molecular interaction variety (nonpolar, electrostatic, pi, etc.), similarity scoring and weighting algorithm, molecule preparation, and presentation of the analysis result. The vastness of the workflow and use case for this methodology could overwhelm the developer of the IFP tool.
Now that we have seen the good and the bad of each end of the spectrum and a glimpse of the complexity of molecular interaction analysis, I suppose it is time for me to explain how a Domain Specific Language could be a better alternative.
First, let’s create an imaginary language to analyse the internal nonpolar interaction inside the protein.
open 1znm.pdb
select "protein" as prot
measure nonpolar in prot
present nonpolar at atomic level
Looks very neat isn’t it? Next, let us specify our atom typing for nonpolar atoms, and then store the result in zinc_finger_nonpolar.txt file.
open 1znm.pdb
select "protein" as prot
specify nonpolar atom with smarts_pattern "[$([CH3X4,CH2X3,CH1X2,F,Cl,Br,I])&!$(**[CH3X4,CH2X3,CH1X2,F,Cl,Br,I])]"
measure nonpolar in prot
present nonpolar at atomic level into zinc_finger_nonpolar.txt
Last, another imaginary language to analyse four different interactions between two molecules:
open 6ma7.pdb
select "protein" as prot
select "resname TPF" as fluconazole
measure nonpolar, hbond, electrostatic, pi between prot and fluconazole
present all at bitstring level into cyp3a4_bitstring.txt
They may seem so simple at first, but what is so incredible and beautiful about this language is how clear and concise yet at the same time, it can incorporate each detail step by step.
And needlessly to say, it is still very simple and may look like a toy. To make it far more powerful we will have to decide the (molecular interaction) domain we are trying to cover. Then we can decide how much control or implementation we could bestow upon our dear user. Something that I will present in the next post.
Revision Note:
- Oct 25, 2023: Add link to ProLIF feature request